EasyML
Module: tutorials.01_EP_single_cell.05_EasyML.run
Section author: Edward Vigmond <edward.vigmond@u-bordeaux.fr>
EasyML
Basics
EasyML was designed to be compatible with C, and C++, and elementary Matlab. You should be able to copy paste equations from these languages and use them more or less as-is in the modeling language. This language is simply a way to describe and markup equations. This language is NOT Turing complete. There is no way to make arrays (but see Arrays), no for loops, no while loops, no functions, and no flow of control. We maybe could add functions later, but they aren’t implemented currently. There are no #ifdefs
C syntax conventions are followed when appropriate. Operator precedence follows C. Every statement ends with ‘;’. C and C++ comments are allowed.
Getting started
To run the translator on a model, simply run
limpet_fe modelname.model
This will produce modelname.c and modelname.h.
From there, you can create a dynamic library by running
make_dynamic_model modelname.c
Upon running successfully, the translator also prints out a list of equations that use functions. We find that the time it takes to solve an ionic model is roughly proportional to the number of function invocations that are called during the main compute routine. limpet_fe counts these invocations so you don’t have to read the generated code to figure out what expressions will be slowing down the computation. Try inserting lookup tables to minimize the number of function invocations.
Defining the math
At its core, the language is made up of 3 types of statements: assignments, declarations, and markup.
Assignments
Assignments look like this:
variable = expression;
These types of statements make up the bulk of the statements within the EasyML language. EasyML differs from C and Matlab in that the order of its statements don’t matter. Variables do not need to be declared before use, and therefore
a = 3;
b = a*a;
CellML is a markup language for describing, amongst other things, ionic models. Many models are published and availmable from the CellML Model Repository. However, this format can only be processed by a machine. CARPentry uses its own MarkUp Language called EasyML.
is the same as:
b = a*a;
a = 3;
A dependency tree is built up from the assignments and as a result,
variables can only be assigned to once.
All variables and numeric constants are floating point numbers.
Right now the parser only recognizes a subset of the C math functions. If you need to add another function email us and let us know.
The absolute time is available with the variable t.
The variable dt is defined and given in ms, but its use is discouraged. Use diff_XXX instead of writing your own update functions if possible. This makes it easy for you to change the integration method later.
If statements
Conditional statements can be done in two ways: C ternary operators or if statements. C ternary operators look like this:
v_abs = (v > 0) ? v : -v;
However, for nested conditionals, entering equations in this format gets tedious and ugly. Instead, we advise that you use this format:
if (v > 0) {
v_abs = v;
} else {
v_abs = -v;
}
This is simply syntaxic sugar for the above ternary form. In general the full syntax for if statements is if () {…} elif () {…} else {…} instead of if () {…} else if () {…} else {…}. C floating point boolean operators are used in as the conditional expressions. The translator recognizes “and, &&, or, ||”.
Note that because the order of operations doesn’t matter, there are some gotchas when using if statements. In particular, these two sections of code are equivalent:
v_abs = -v;
if (v > 0) {
v_abs = v;
}
versus
if (v > 0) {
v_abs = v;
} else {
v_abs = -v;
}
To understand what value a variable takes in the presence of multiple branches, you can read the code top to bottom. This should let you copy and paste code from your existing models.
However, because of this quirk, you can’t do things like this that you could do in C
A = 50*x*x+40*x+100;
if (A > 2000) {
A = 2000;
}
This does not clamp the variable above 2000 in the way you expect, because this is equivalent to
A = (A > 2000) ? 2000 : 50*x*x+40*x+100;
which means A depends on itself, which in turn means that A will have to be a state variable. This is not what you want. Rewrite this equation to
A_factor = 50*x*x+40*x+100;
A = A_factor;
if (A_factor > 2000) {
A = 2000;
}
Variable updates
We also have syntaxic sugar for accumulating variables. The following two codes are equivalent: CellML is a markup language for describing, amongst other things, ionic models. Many models are published and availmable from the CellML Model Repository. However, this format can only be processed by a machine. CARPentry uses its own MarkUp Language called EasyML.
Iion = 0;
Iion += INa;
Iion += Ito;
versus
Iion = 0+INa+Ito;
Markup
Markup statements change the way the code handles some variables compared to others. A markup statement takes the form:
.markup_function(optional_arguments);
The markup function is applied to the variable definition or declaration immediately preceding the markup command. Therefore,
X; .external();
Y = 3;
Z = 3; .external();
applies the .external markup to X and Z. If a markup follows an if statement, the markup applies to every variable definition or declaration enclosed in that if statement.
In order to avoid tedious repetition of markup statements, the group directive can be used:
group {
X;
Z = 3;
} .external();
Y = 3;
group statements can be nested, and like an if statement, a group statement collects every declared/defined variable it encloses for markup.
Recognized Markup
Right now, here are the markups that the limpet_fe translator understands / respects :
| markup | meaning |
|---|---|
| .external([name]) | the variable can be used externally to the model in the LIMPET code. An optional argument lets you specify the external name if different from the variable’s name. |
| .flag(A,B,…) | The variable can only take on the values specified in the argument,
and is set with the flags parameter. |
| .lb() | clamped lower bound for variable |
| .lookup(min,max,step) | Define a lookup table on this variable with lower bound min, upper bound max, and with step as the discretization of the table. |
| .method(integration) | integration method for equation. Available values are:
|
| .nodal() | parameter varies on a node by node basis. If used with .external, the variable needs to be read/written to using the GlobalData_t arrays. If not used with .external, the parameter becomes a state variable even if the compute function never changes its value. |
| .param() | Marks a variable as being a parameter that the user should be able to adjust from the command line. |
| .regional() | variable is constant on a per-region basis, and if used with .external, means the variable can be found in the cgeom structure. |
| .store() | Manually written update function which ignores the differential update. Take this example from RDII_F and how it implements the LRd 2000 Zeng dynamics for Calcium: dCa_i = -((i_Cai*acap)/(vmyo*zCa*F) +i_up *vnsr/vmyo -irelcicr *vjsr/vmyo-
ireljsrol*vjsr/vmyo)*dt;//differential equation is ignored
Catotal = trpn+cmdn+dCa_i+Ca_i;
bmyo = cmdnbar+trpnbar+kmtrpn+kmcmdn-catotal;
cmyo = kmcmdn*kmtrpn-catotal*(kmtrpn+kmcmdn)+trpnbar*kmcmdn+cmdnbar*kmtrpn;
dmyo = -kmtrpn*kmcmdn*catotal;
fmyo = sqrt(bmyo*bmyo-3*cmyo);
Ca_i = ((2*fmyo/3)*cos(acos((9*bmyo*cmyo-2*bmyo*bmyo*bmyo-27*dmyo)/
(2*pow((bmyo*bmyo-3*cmyo),1.5)))/3)-(bmyo/3));.store();//analytical solution
This formulation for Calcium is preferred for LRDII_F because it avoids extremely stiff differential equations for the Troponin calcium buffers. |
| .trace() | Says that a column for this variable should be written when defining a trace_XXX() function. |
| .ub() | clamped upper bound for variable |
| .units(unit_expression) | units of variable. Units can be written as arbitrary unit expressions, and can use all SI prefixes (pnumckMG). Recognized units are:
|
Declarations
Declarations look like this:
variable;
These do nothing, but are useful for applying markup.
Special variable name rules
Some variables have special meanings. Here are the special meanings that the translator currently recognizes:
| variable | meaning |
|---|---|
| diff_XXX | the differential equation for variable XXX. All derivatives should be with respect to the same time unit. If no units are specified, then milliseconds is assumed. |
| d_XXX_dt | Same as diff_XXX |
| XXX_init | initial value for XXX |
Gates
The equations for a Hodgkin-Huxley type gate are best described by:
| variable | meaning |
|---|---|
| alpha_XXX or a_XXX | the opening 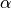 rate rate |
| b_XXX or beta_XXX | the closing 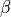 rate rate |
| tau_XXX | the time constant of change |
| XXX_inf | the steady state value |
In order for a variable to be marked as a gate, then XXX must be used as a variable in another equation AND one of the pairs (alpha, beta) or (tau, inf) must be found. When a variable is marked as a gate, a diff_XXX equation will be added for it automatically. The variable will be solved using Rush Larsen, and an XXX_init variable will be defined if none exists.
Note
Defining 3 or more of these variables is redundant and can cause problems. Do NOT define diff_XXX or d_XXX as they will automatically be generated
States
States are like gates but their change is not described by a differential equation. They may, for instance, discretely
change value based on voltage and time. To define a state variable, make sure to initialize it as above, and when the value is updated, use the .store(); markup. For example, to use a variable called activated:
activated_init = 0;
if( activated==0 and V>-20 ){
new_act = 1;
}elif( activated==1 and V<-20 ) {
new_act = 2;
} else {
new_act = 0;
}
activated = new_act;.store();
Arrays
Arrayed values may be particularly useful when defining compartmentalized models with repeated sections. Variable arrays are not supported in EasyML but preprocessing with arrayifier.py unrolls array variable expressions to mimic their fuctionality:
arrayifier.py -h
usage: arrayifier.py [-h] [--def-size DEF_SIZE] [--index INDEX] [--process]
[--limpet-fe LIMPET_FE]
arrayed [unrolled]
fake arrays in limpet_fe
positional arguments:
arrayed original model file with array syntax
unrolled output model (stdout)
optional arguments:
-h, --help show this help message and exit
--def-size DEF_SIZE default array size (10)
--index INDEX loop index in RHS expr (__INDEX__)
--process run limpet_fe on unrolled file
--limpet-fe LIMPET_FE
limpet_fe program (limpet_fe.py)
A new markup function .array() has been added as well as an array syntax.
Let V be a variable. To make it an array of N values
V;.array(N);
group {V1;V2;}.array(N);
V;.array();
With no size specified, a default is used which can be changed as an option (–def-size) to arrayifier.py.
To assign to variable slices, use python slice syntax V[a:b:c] which is the set of values [a,a+c,a+2*c,…,b-1] A missing “a” means a=0, and omitting “b” implies N, and negative numbers are measured from the end Note, an extension is added: a leading ^ means take the complement of the slice specified. Eg, given V has 6 entries V[^1:3] => V[0],V[3],V[4],V[5] To assign to a particular index
V[2] = ...
The RHS may also contain array variables which must match in size the LHS. All slices must be the same size. Be careful not to assign to an entry which has been previously assigned in a slice. The following is a 1D diffusion example
diff_V[1:-1] = k*(V[2:]+V[:-2]-2*V[1:-1])
Warning
Slice specifications cannot be expressions. eg [a+1:] and [1-1:5] are INVALID.
__INDEX__ is recognized as the enumeration of the LHS set, eg
V[1:] = 2+__INDEX__;
would lead to
V[1] = 2;
V[2] = 3;
V[3] = 4;
Another word can be chosen with a commandline option
If the initial predicate of the if statement contains an array, the whole expression is unrolled including elif’s and else’s, eg
if( a[:] > 3 ) { b[:] = 2; } elif( a[:] < 1 ) {b[:] = 2; } else { b[:] = __INDEX__ }
becomes
if( a[0] > 3 ) { b[0] = 2; } elif( a[0] < 1 ) {b[0] = 2; } else { b[0] = 0 }
if( a[1] > 3 ) { b[1] = 2; } elif( a[1] < 1 ) {b[1] = 2; } else { b[1] = 1 }
etc
If the predicate does not contain an array, arrays are expended as needed, eg
if( foo == 1 ) {bar[:] = 3; }
becomes
if( foo==1 ){ bar[0] = 3; bar[1] = 3; bar[2] = 3; ... }
elif predicates can contain arrays if and only if the initial predicate does. For nested if’s, the outermost arrayed initial predicate determines the array index of any contained array expressions.
Derived variables are automatically arrayed if the original variable is:
diff_V[:] = f(V[:],Ca[:])
alpha_X[:] = ...
b_X[:] = ...
tau_X[:] = ...
X[:]_inf = ...
To initialize variables:
V[:]_init = ...
V[2]_init = ...