Limit cycle initialization
Module: tutorials.02_EP_tissue.03B_study_prep_init.run
Section author: Fernando Campos <fernando.campos@kcl.ac.uk>
This tutorial demonstrates how to initialize a cardiac tissue with state variables obtained from a single-cell stimulation.
Conceptual Overview
Similar to in-vitro experiments, cardiac tissue is usually stabilized by pacing it at basic cycle
length (BCL) in order to reduce beat-to-beat changes. However, stabilization might require a few
hundred beats which can take days or even weeks to complete in organ-scale models. Unlike tissue
simulations, pacing an isolated cell until it achieves its stable cycle limit is computationally
feasible. In order to save computational efforts, the single-cell model state variables ( ,
channel gating variables, etc.) at the end of the pacing protocol can be stored and used to initialize a
tissue model. This initialization procedure is equivalent to pacing the entire tissue model in a
space-clamped mode. Although this procedure is computationally cheap, it is biologically unrealistic
since it ignores any differences due to the activation sequence of a previous beat. If information
on the activation sequence in the cardiac tissue is available before hand, it can be used in combination
with the single-cell initialization method to overcome this limitation.
,
channel gating variables, etc.) at the end of the pacing protocol can be stored and used to initialize a
tissue model. This initialization procedure is equivalent to pacing the entire tissue model in a
space-clamped mode. Although this procedure is computationally cheap, it is biologically unrealistic
since it ignores any differences due to the activation sequence of a previous beat. If information
on the activation sequence in the cardiac tissue is available before hand, it can be used in combination
with the single-cell initialization method to overcome this limitation.
Problem Setup
This example will run a simulation using a 2D sheet model (1 cm x 1 cm) in which all cells are
initialized with single-cell model states stored in file INIT.sv. The file contains initial
conditions of all ordinary differential equations (ODEs) describing channel gating and ionic
concentrations in the chosen cellular model. The initial conditions are obtained after pacing
the cell model at a prescribed BCL for a number N of beats.
Usage
To run this tutorial :
cd tutorials/03B_study_prep_init
./run.py --help
The following optional arguments are available (default values are indicated):
--model Model of the cellular action potential
Options: {MBRDR,TT2}
Default: Modified Beeler-Router Drouhard-Roberge (MBRDR) model
--init-method Method to initialize the tissue model
Options: {none, sv_init or prepace}
Default: none
--bcl Basic cycle length for repetitive single-cell stimulation
Default: 500 ms (2 Hz)
--npls Number of pulses applied to the single-cell with the given bcl
Default: 10
If the program is run with the --visualize option, meshalyzer will automatically load transmembrane
potentials  :
:
./run.py --visualize
Key Parameters
The key parameter is --init-method. This will be set to one of three possible values in order
to initialize the 2D sheet model:
- none: all cells are initialized with the cellular model default states that are hard coded in CARP
- sv_init: all cells are initialized with the cellular model states stored in file
INIT.sv. Model states are computed here by running a single-cell simulation, where the cell is pacednpls(default: 10 pulses) times with the prescribedbcl(default: 500ms). Model states are taken at the end of the simulation protocol. - prepace: unlike in the sv_init initialization method, where state variables are saved into a file at a prescribed time instant (usually at the end of a single-cell pacing protocol), in the prepace method cells are initialized with model states taken at different time instants depending on their a priori known activation time. The difference between the two methods is illustrated below:
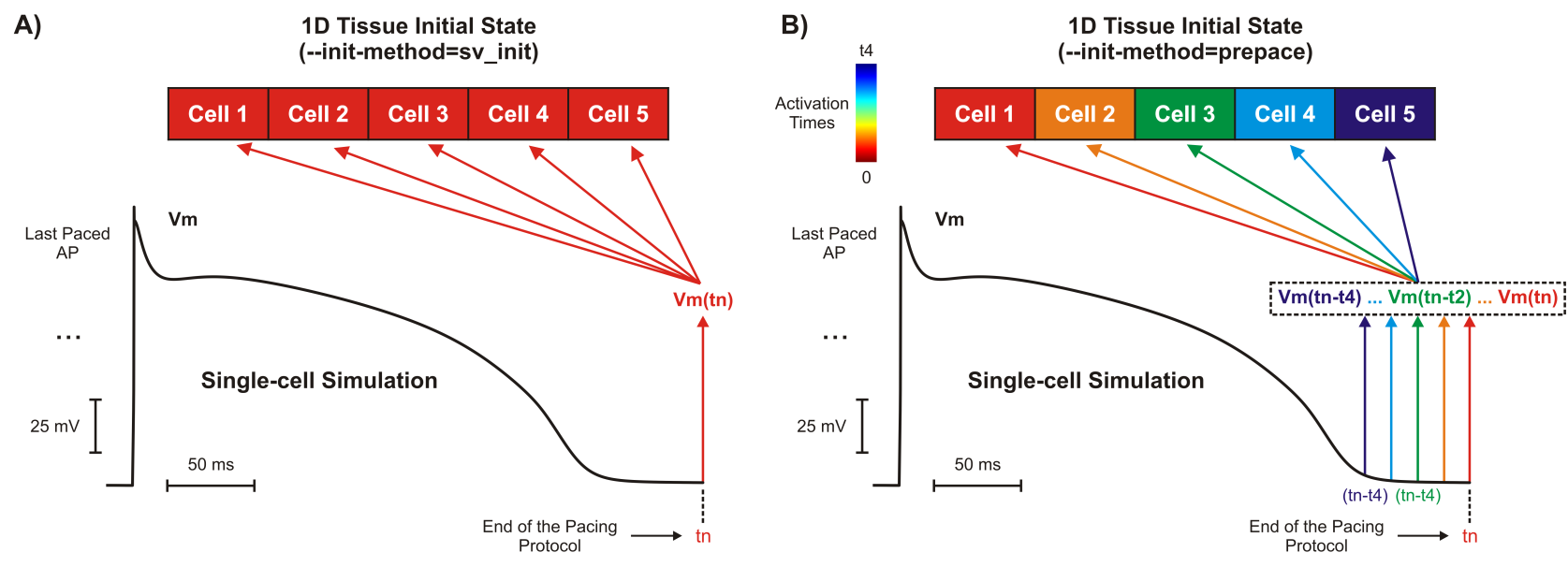
Fig. 70 Tissue initialization methods. A) A 1D tissue model with all cells initialized with the same
 as well as other model state variables (not shown) taken at the end of the
single-cell simulation. B) The same 1D tissue as in A) but cells are initialized with
as well as other model state variables (not shown) taken at the end of the
single-cell simulation. B) The same 1D tissue as in A) but cells are initialized with  and other state variables (not shown) taken at different time instants. The time instants are
determined based on the activation sequence know before hand: cell 1 activates at 0, cell 2 at
t1, …, and finally cell 5 at time t4.
and other state variables (not shown) taken at different time instants. The time instants are
determined based on the activation sequence know before hand: cell 1 activates at 0, cell 2 at
t1, …, and finally cell 5 at time t4.
Interpreting Results
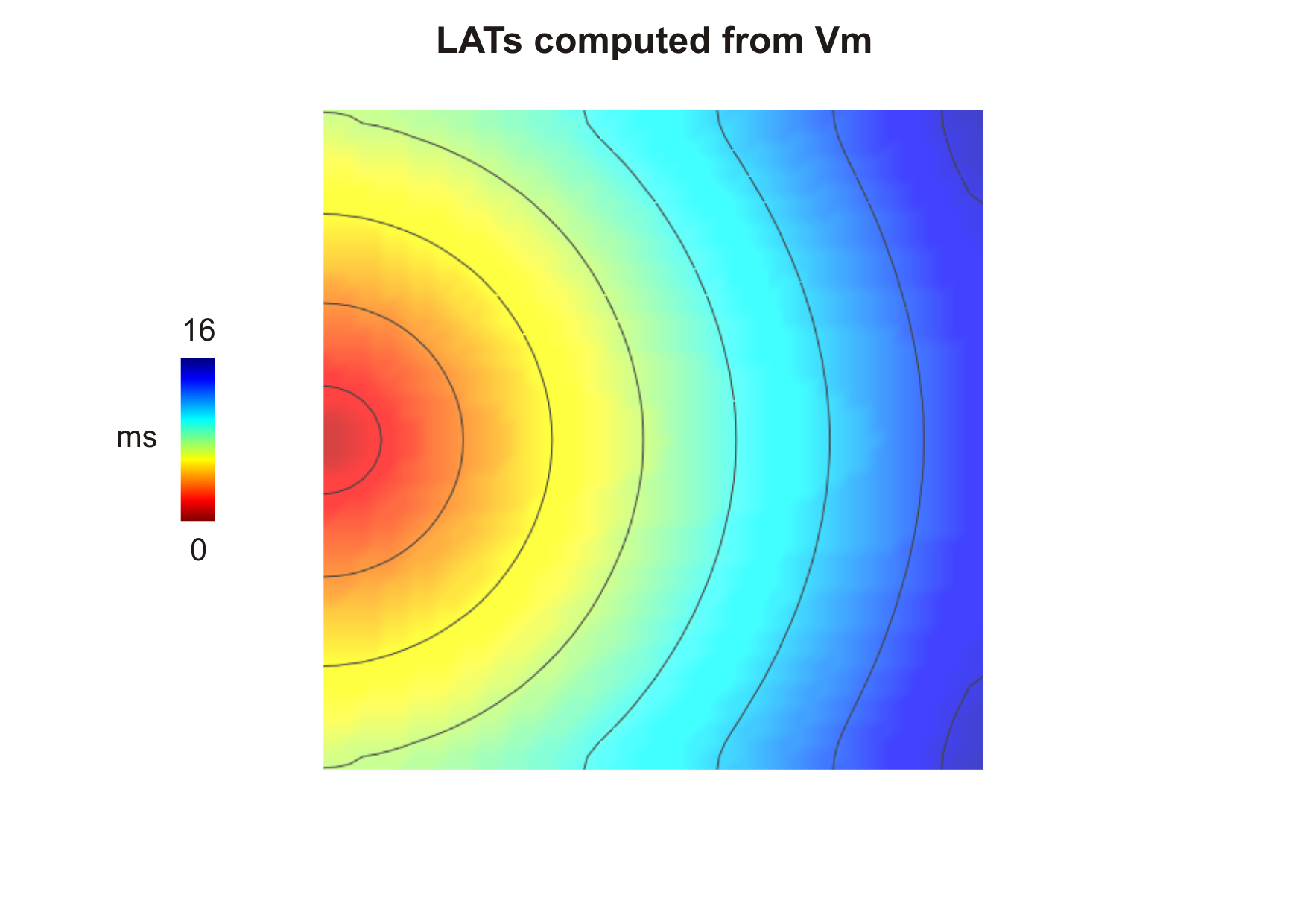
The difference between the results produced with each initialization option (i.e.: none, sv_init and
prepace) can be appreciated by looking at the spatial distribution of  at time t = 0 with meshalyzer
(
at time t = 0 with meshalyzer
(--visualize):
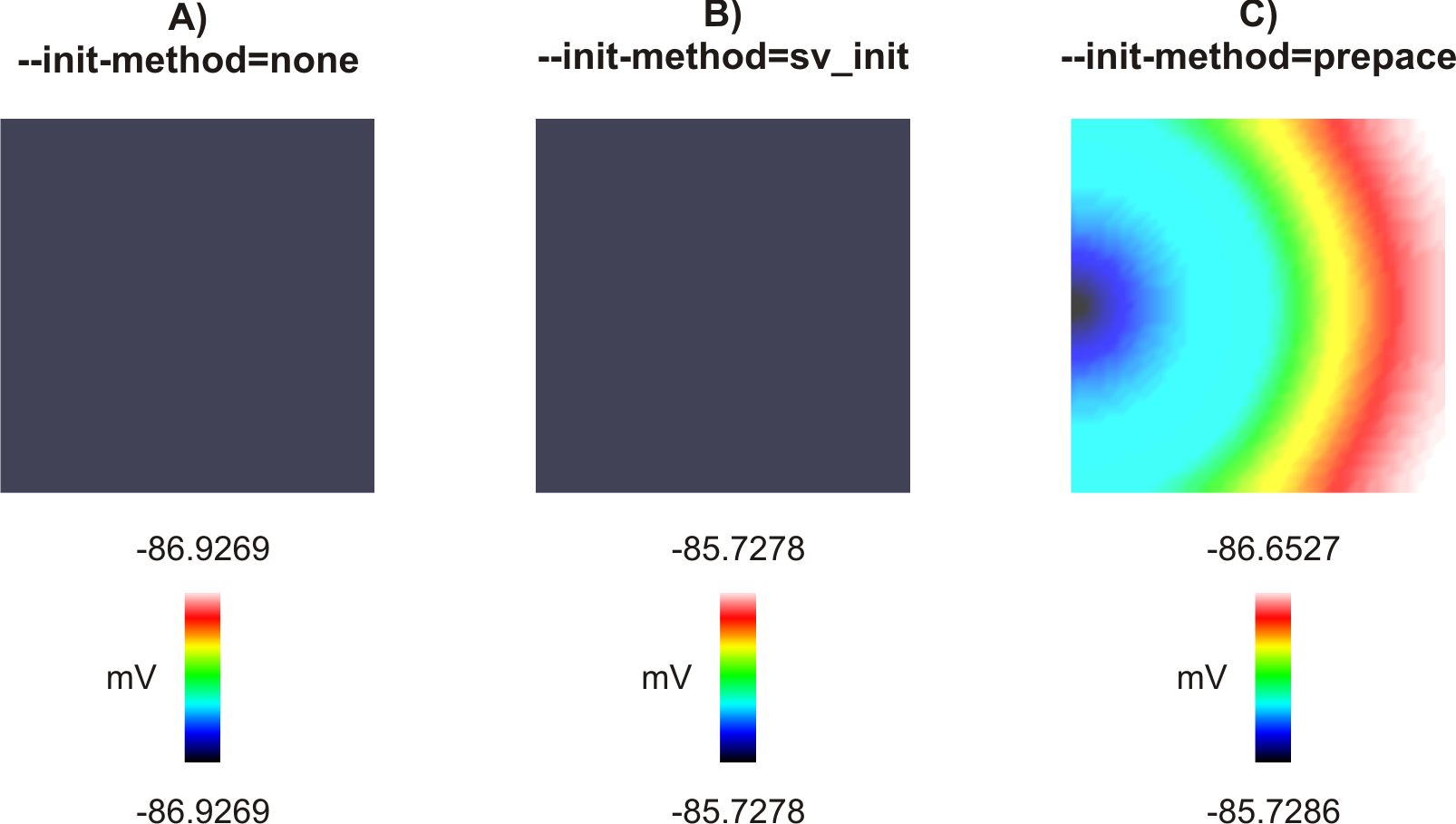
Fig. 71 Spatial distribution of  at the beggining of the simulation (t = 0 ms). A) All cells in the
tissue are initialized with default model states of the MBRDR cell model. B) All cells in the tissue are initialized
with the same model states in file
at the beggining of the simulation (t = 0 ms). A) All cells in the
tissue are initialized with default model states of the MBRDR cell model. B) All cells in the tissue are initialized
with the same model states in file INIT.sv. the same model states in file INIT.sv. C) Cells in the tissue
are initialized with model states taken at different time instants depending on their a priori known activation time.
- A) By default (
--init-method=none), at t = 0 ms will be -86.9269 mV for all cells in the tissue.
This is because -86.9269 is the default initial condition of
at t = 0 ms will be -86.9269 mV for all cells in the tissue.
This is because -86.9269 is the default initial condition of  in the
in the MBRDRmodel (the initial condition of in the TT2 is -86.2).
in the TT2 is -86.2). - B) By running with the
--init-method=sv_initoption, at t = 0 ms will be -85.7278 mV for all
cells in the tissue. This is because -85.7278 is the initial condition of
at t = 0 ms will be -85.7278 mV for all
cells in the tissue. This is because -85.7278 is the initial condition of  in file
in file INIT.sv, all model states were obtained after pacing theMBRDRmodel for 10 times with a bcl of 500 ms. - C) By running with the
--init-method=prepaceoption, at t = 0 ms will vary between -85.7286 mV
(left lower corner) and -85.6527 mV (right upper corner). This is because, unlike the previous
methods where all cells are initialized with the same cellular model states, cells are initialized
according with model states obtained at different time instants (see above) according to the
activation sequence in file
at t = 0 ms will vary between -85.7286 mV
(left lower corner) and -85.6527 mV (right upper corner). This is because, unlike the previous
methods where all cells are initialized with the same cellular model states, cells are initialized
according with model states obtained at different time instants (see above) according to the
activation sequence in file LATs.dat:
Although the differences in  between the three initialization methods seem negligible,
they can be substantial in other state variables of the ionic model with a slower time decay such as the
intracellular calcium
between the three initialization methods seem negligible,
they can be substantial in other state variables of the ionic model with a slower time decay such as the
intracellular calcium 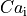 .
. 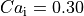 if
if --init-method=none and
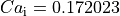 (see file
(see file INIT.sv) if --init-method=sv_init with the bcl = 500 ms
and npls = 10. These differences increase for all state variables as the bcl is reduced.
CARPentry Parameters
The relevant parameters used in the tissue-scale simulation are shown below:
# Model of the cellular action potential
-imp_region[0].im = model
# Used by the sv_init method
-imp_region[0].im_sv_init = 'INIT.sv'
# Used by the prepace method
-prepacing_beats = 10
-prepacing_lats = 'LATs.dat'
-prepacing_bcl = 500
imp_region[] is used to assign a model of cellular action potential to all cells in the tissue. If
the sv_init initialization method is choosen, then the tissue will be initialized with cellular
model states in file INIT.sv. However, if the prepace method is choosen, then the cell model
in imp_region[] will be paced npls times with the given bcl. Tissue activation times in file
LATs.dat are used to guide state set-up for prepacing.
Once the initial conditions are prescribed, the tissue simulation is started. In this example, a transmembrane current of 100 uA/cm^2 is applied for a period of 1 ms to the cells located at the middle of the left side of the tissue.