Adjusting action potential duration
Module: tutorials.02_EP_tissue.03_study_prep_APD.run
Section author: Jason Bayer <jason.bayer@ihu-liryc.fr>
This tutorial demonstrates how to adjust ionic model parameters to generate a specific action potential duration in your simulations.
Introduction
In mammalian hearts, cardiac repolarization varies between species, is spatially heterogeneous, and altered during disease. Thus, it is important to calibrate your ionic models to account for these repolarization differences. The easiest way to do this is to modify the maximum conductance for ionic currents known to alter action potential duration (APD), i.e. the time duration a cardiac myocyte repolarizes after excitation. Ideally, these values are obtained from experiments in the literature. In this exercise, you will learn how to look up the ionic current variables in your model that you can change to alter APD, and how to adjust in cable/tissue models.
Problem setup
To execute simulations of this tutorial do
cd tutorials/03_study_prep_APD
Ionic model
For this example, we use the most recent version of the ten Tusscher ionic model for human ventricular myocytes [#Tusscher2006]. This ionic model is labeled TT2 in CARP’s LIMPET library.
1D cable model
For this exercise we compute APD in a 1.5 cm cable of epicardial ventricular myocytes. The model domain was discretized with linear finite elements with an average edge length of 0.02 cm.
Pacing protocol
The left side of the 1D cable model is paced with 5-ms-long stimuli at twice capture amplitude for a basic cycle length and number of beats chosen by the user.
Action potential duration
Activation potential duration is computed at 80% repolarization (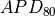 ) according to [Bayer2016]. This is achieved by using the igbutils function igbapd as illustrated below. See the igbutils tutorial for more information on using igbapd.
) according to [Bayer2016]. This is achieved by using the igbutils function igbapd as illustrated below. See the igbutils tutorial for more information on using igbapd.
./igbapd -t nbeats-1*bcl --repol=80 --vup=-10 --peak-value=plateau --plateau-start=15 ./vm.igb
Usage
The following optional arguments are available (default values are indicated):
./run.py --help
--Mode Options: {default,adjust}, Default: default
When in adjust mode, will use im_param options by user
--im_param Is a string for which you can input operations to
perform to specific variables in a chosen ionic model.
For example, 'Gmax_1*0.4,Gmax_2=0.4'
--IMP Ionic model name from LIMPET library.
--nbeats Default: 2
Number of beats to perform before APD computation. In
reality, this should be much larger to reach
steady-state. Due to tutorial time constraints, it is
very small.
--bcl Default: 700
Basic cycle length of pacing
Tasks
- List all of the ionic channel conductances in the ten tusscher model by running the command below
bench --imp-info -I TT2
- Determine the baseline APD by running the following command.
./run.py --Mode default --IMP TT2
Note, you can look at the values for the APD on each node of the cable by scrolling through the vertex values under the Highlight tab in meshalyzer.
- Shorten APD by >30 milliseconds by adjusting the maximal conductance for the slow outward potassium current (Iks). What is the name of the variable from the output in task 1? Did you have to increase or decrease the value?
- This time, prolong APD by >30 altering the plateau of the action potential by modifying the maximal conductance of the L-type calcium channel current (ICaL).
In the future, you will want to run many more beats than 2. Also, the process can be automated similar to tuneCV.
References
| [1] | ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol, 291(3):H1088-H1100, 2006. [Pubmed] |