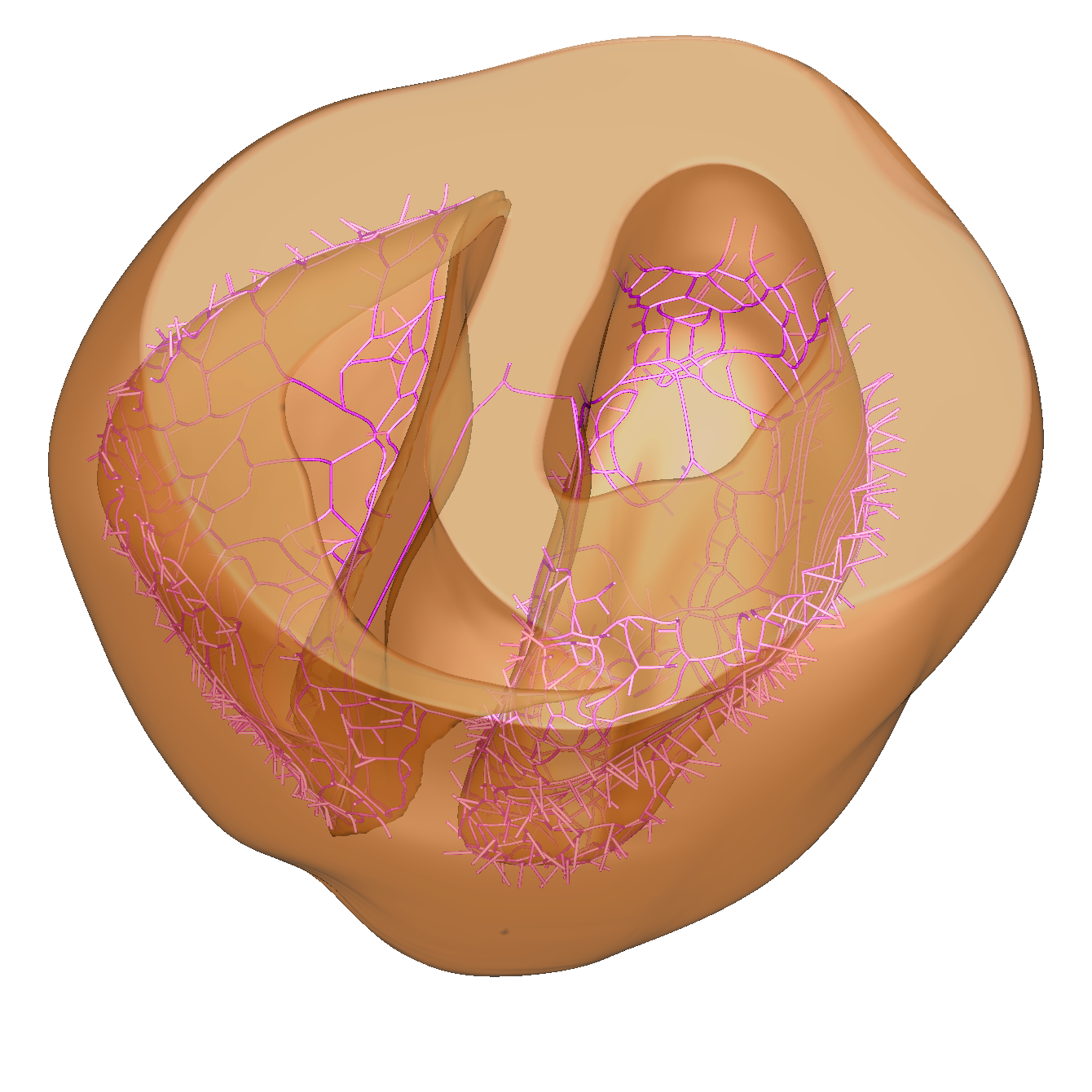
Purkinje System
Module: tutorials.02_EP_tissue.19_Pkje.run
Section author: Edward Vigmond <edward.vigmond@u-bordeaux.fr>
Purkinje System
Section author: Edward Vigmond
The Purkinje System (PS) is the fast conducion system of the ventricles. It starts with the bundle of His, and then bifurcates into left and right bundle branches. The left bundle branch (LBB) splits into three fascicles, while the Right Bundle Branch (RBB) has a branch which follows the moderator band.
Connections are only made at discrete end points, called Purkinje-Myocyte Junctions (PMJs) with anterograde conduction across the PMJ being much slower (3-12 ms) than retrograde conduction (1-5 ms). This asymmetry in conduction can be captured.
Specification
The file describing the PS is separate from the finite element mesh. It has a specific format as described in Purkinje file. Each cable in the system is allowed to have up to 2 cables which branch from it, the sons. Simialrly, a cable can have up to 2 parent cables.
Nodes in the PS are defined in the PS file so they do not have to be nodes of the ventricular finite element mesh.
Purkinje-Myocyte Coupling
PMJs are formed at the end of a cable with no sons.
For propagation to occur from the PS to the myocardium, a liminal volume must be stimulated, and not just a single myocardial node.
All myocardial nodes less than PMJsize micrometers from the
terminal node in the PS cable are coupled to the
Purkinje node.
If at least nPMJmin nodes are not contained within the PMJ volume,
or more than nPMJmax nodes are contained,
PMJsize is automatically adjusted until the conditions are satisfied.
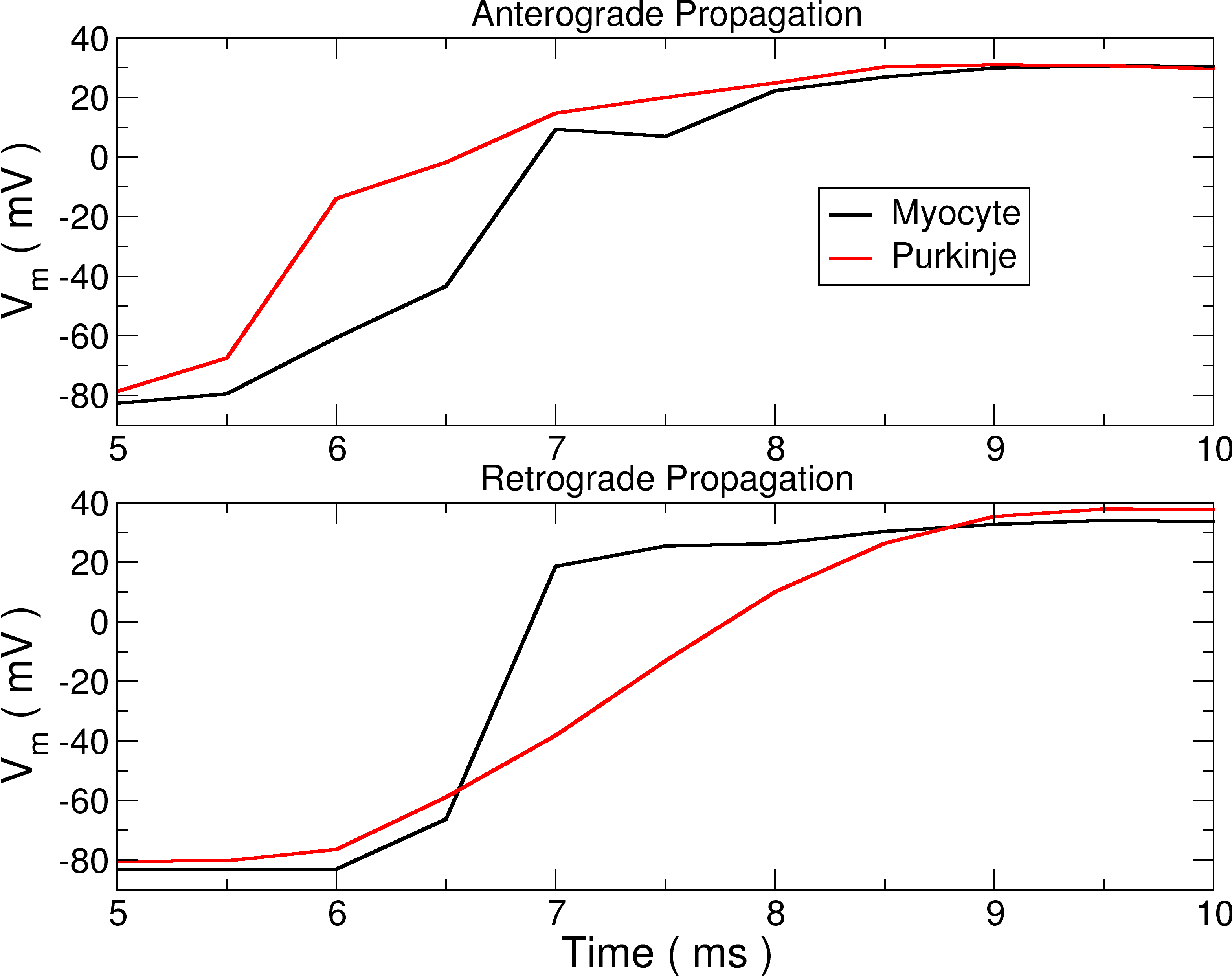
Fig. 119 Asymmetry in propagation across a PMJ. The top trace shows activity entering from the PS into the myocardium, while the bottom shows activity from the myocardium entering the PS.
Anterograde propagation, from the PS to the myocardium, is described by
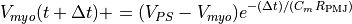
with 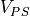 and
and 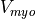 being
the PS and myocardial transmembrane voltages respectively.
The PS is assumed to branch within the PMJ volume, below the level of discretization, so that the load seen is reduced by the factor
being
the PS and myocardial transmembrane voltages respectively.
The PS is assumed to branch within the PMJ volume, below the level of discretization, so that the load seen is reduced by the factor PMJscale.

By adjusting Rpmj and PMJscale, the asymmetry of conduction across the PMJ can be tuned.
| Option | Meaning |
|---|---|
| Rpmj | the resistance of PMJ juntion |
| PMJscale | divide PS load by this factor |
| nPMJ_min | minimum number of myocardial nodes |
| nPMJ-max | maximum number of myocardial nodes |
| PMJsize | distance from end point to search for myocardial nodes |
Propagation
Propagation velocity within the PS is rapid, >4 m/s.
The is controlled by setting the values of the sodium current in the ionic model,
and the values of the conductivity (purk_cond) and gap junction resistance
(purk_resist) in the PS.
Conductivity is homogenized, 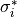 , and that value used as defined below.
, and that value used as defined below.
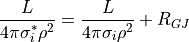
if purk_effcond is nonzero, the value of cell_length is used for the value of
L, else, the actual internodal distance is used. If purkIgnoreGlobals
is nonzero, then the values of resistance and conductivity are set on a cable
by cable basis as defined in the .pkje file,
else they are set globally from the command line options below.
| Option | Meaning |
|---|---|
| purk_effcond | effective conductivity is constant, else cable specific |
| purk_cond | intracellular conductivity of all PS cables, 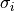 |
| purk_resist | gap junction resistance between cable, 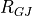 |
| purkIgnoreGlobals | ignore purk_resist and purk_cond if set |
| cell_length | nominal cell length |
Experimental Setup
Experiments will be run in a slab with a relatively simple Purkinje network composed of a His cable
Experiments
The below defined experiments demonstrate propagation from a single cable into a piece of tissue. To run these experiments
cd tutorials/02_EP_tissue/19_Pkje
Run
./run.py --help
to see all exposed experimental parameters
--Rpmj RPMJ resistance across PMJ (1000.0 kOhm)
--cond COND PS conductivity (0.6 S/m)
--Rgj RGJ resistance between PS cells (100.0 kOhm)
--GNa GNA scale conductance of sodium channel (1.0)
--PMJscale PMJSCALE scale PS current load (1000.0)
--PS-act {His,retro} type of Purkinje system activation (His)
--nPMJ NPMJ NPMJ number of min and max myocardial nodes/PMJ [10, 30]
Experiment exp01
Observe transmission across the junction in the anterograde direction, that is, from the PS into the myocardium.
./run.py --visualize
How does it change if you change the value of Rpmj or PMJscale?
./run.py --visualize --Rpmj=10000
Experiment exp02
We will look at retrograde conduction which occurs when activity from the myocardium enters into the PS by crossing the PMJ.
./run.py --visualize --PS-act=retro --PMJscale 1000
How does it change if you change the value of the junctional parameters like Rpmj or PMJscale?
Literature
| [1] | Clements and Vigmond, Construction of a computer model to investigate sawtooth effects in the Purkinje system, IEEE Trans Biomed Eng, 54(3):389-99, 2007. [PubMed] [Full] |
| [2] | Boyle, Deo, Plank, and Vigmond, Purkinje-mediated effects in the response of quiescent ventricles to defibrillation shocks, Ann Biomed Eng, 38(2):456-68, 2010. [PubMed] [Full] |