Forward modeling
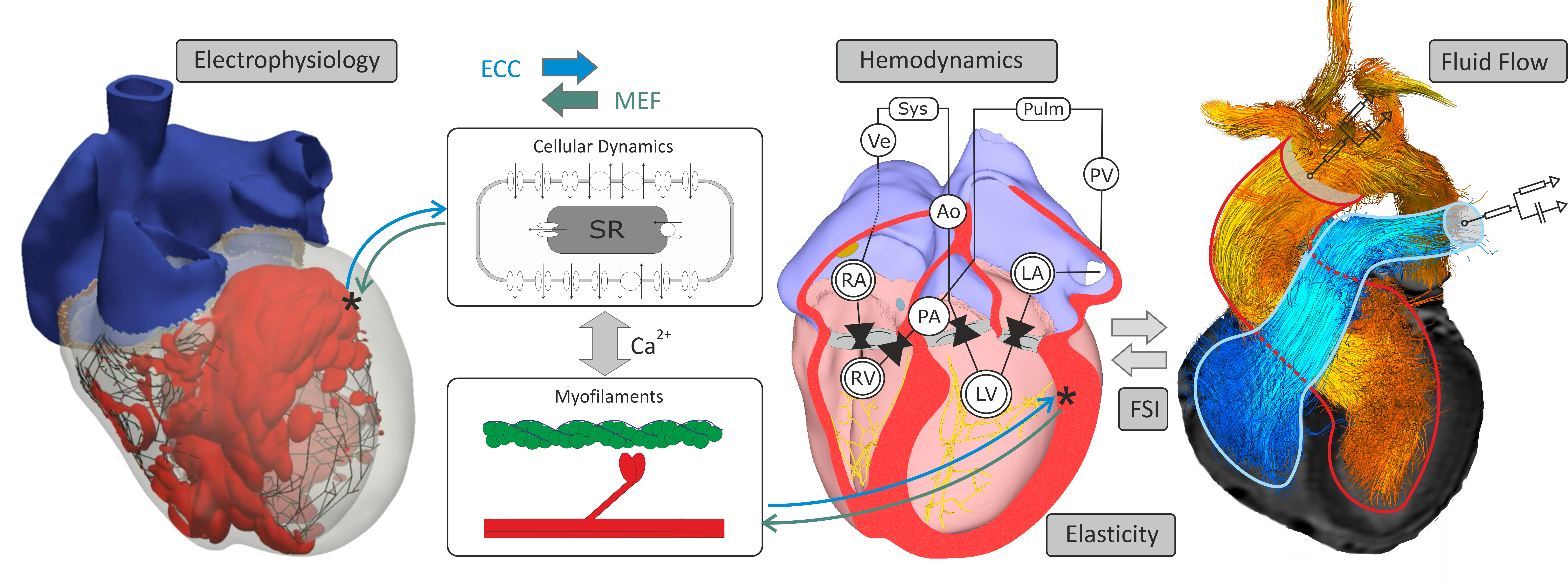
Fig. 32 Cardiac function
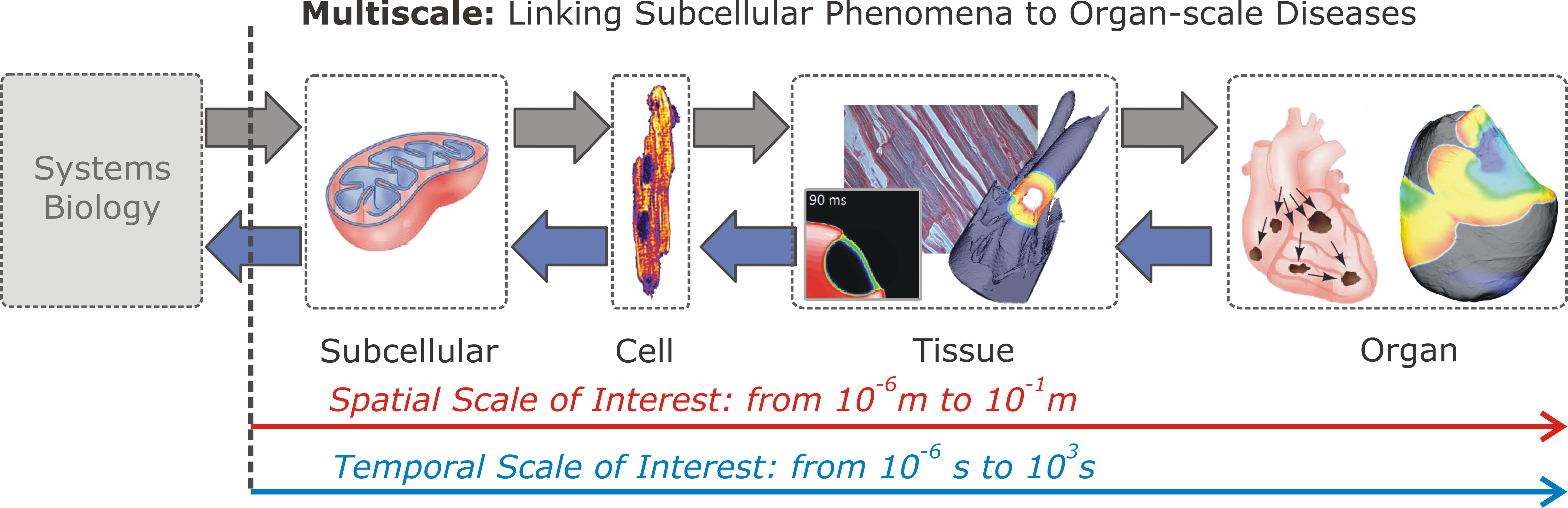
Fig. 33 Multiscale aspects of cardiac function
Model Parameterization and Initialization
While generic models using generic parameter sets and initial values can be valuable and provide generic mechanistic insights, other applications prompt for using specific parameter sets and initial values to facilitate model-based predictions on a case-by-case basis. In such application such as the use of modeling in the clinic as a tool for planning and optimizing therapies, stratification of patients and the prediction of outcomes it is necessary to personalize models, that is, to tune the model behaviour for a specific individual. Within the equations presented, many parameters are required which are based on physiology. However, getting accurate estimates of these is challenging for many reasons as discussed below:
- Anatomical variability: The heart is an organ of high plasticity which changes its anatomical shape over a lifetime as a function of the imposed workload. If a heart is overloaded by either volume or pressure to cause stresses or strains within the myocardium above a critical threshold, remodelling processes are triggered leading to concentric or eccentric shape changes of the ventricles. Apart from pathological alterations in shape due to congenital malformations or pathological overload conditions, cardiac anatomy varies significantly not only between species, but also between individuals of the same species. Clinical modeling applications attempt to represent an individual’s heart anatomy based on tomographic imaging data as accurately as possible. For such applications specialized model building workflows have been developed which seek to build anatomical models of a given patient’s heart as automated as possible to minimize the need for operator interventions (See Fig. 4).
- Spatial heterogeneity: Cells naturally possess differences with no two cells perfectly equal. Shape and protein expression differ between cells in the same region, due to the slightly different environments seen and the stochastic nature of cellular processes. Across larger spatial scales, clear gradients are observed, as from the apex to the base, from the endocardium to the epicardium, and between the left and right ventricles. Furthermore, protein expression levels also follow a circadian rhythm which can be significant, so sample acquisition time is a factor.
- Incompleteness of data: A complete data set to fully parameterize a cannot be obtained from a single sample. Many samples are require to confidently measure a particular property or quantity, and there are many quantities to measure. Reported values are averages, but given the nonlinear behaviour of biological systems, there is no guarantee that using averaged properties result in typical behaviour.
- Sample preparation: Experiments to determine parameter values require disruption of the natural state of the heart. Even with in tact hearts, function is severely affected. Simply opening the torso affects thoracic pressure with consequences. Excising a heart, i.e. a Langendorff set up, removes the influence of circulating hormones, and neurotransmitters, as well as eliminates central nervous input. Furthermore, performance degrades over time as the blood supply is disrupted leading to reduced energy stores and accumulation of waste products. Tissue preparations go further in removing mechanical load. With single cell preparations, the isolation procedure involves digestion of the interstitial collagen which also changes cell shape. Protein turnover of membrane channels is highly dynamic, over a time scale of minutes, and isolation affects ion channel expression.
- Tissue structure: The rotating transmural predominant fibre orientation makes direct measurements of material quantities impossible since the properties are anisotropic and dictated by cellular direction which change significantly over the distance of microns. A fibre orientation distribution must be assumed. For example, one cannot measure conductivity values by simply measuring the voltage associated with an applied current. Estimates of electrical conductivities are bounded to a maximum of 1 S/m but vary widely.
- Clinical Data: For modelling human hearts, clinical data is constrained since measurements cannot be destructive, are limited in time, and tend to be reflect function at the most macroscopic scale. Cadaver hearts are used but their availability is limited, they are usually diseased since they are unsuitable for transplantation, and the pathologies presenting vary widely leading to weak statistical significance. As a result, and not limited to just human models, missing data is supplanted with data from other species which may differ significantly.
- Spatiotemporal scales: Processes in the cell occur at many different time scales, some of which are too fast to accurately measure. Other events may take place over days. Within a cell, important events may occur in nanodomains which are missed if sampled too coarsely.
- Nonspecific and indirect interactions: Drugs or other manipulations are performed to determine parameter values. However, the action may affect more than what is intended. For example, a drug intended to block a particular channel may also affect other channels. Likewise, given the complex feedback systems in biology, a manipulation to one entity may lead to several downstream effects.