Lead field validation benchmark
Module: benchmarks.bidomain.TBunnyC2.run_lf
Section author: Gernot Plank <gernot.plank@medunigraz.at>
EGM computation based on a lead field approach
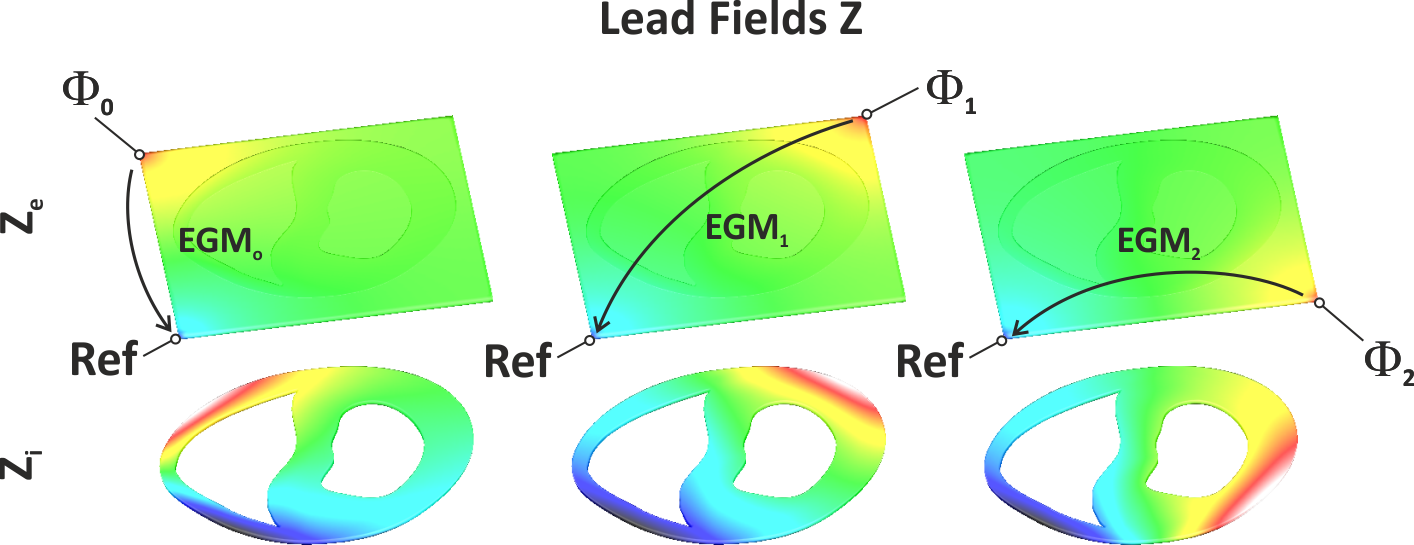
Fig. 29 Shown are the lead fields Z for three EGMs. In all cases the reference electrode is located at the lower left corner of the bath. Electrodes for computing buoikar EGNs relative to the chosen reference are located at the lower right corner :math:’Phi_0’, the upper left corner, :math:’Phi_1’ and the upper right corner, :math:’Phi_2’.
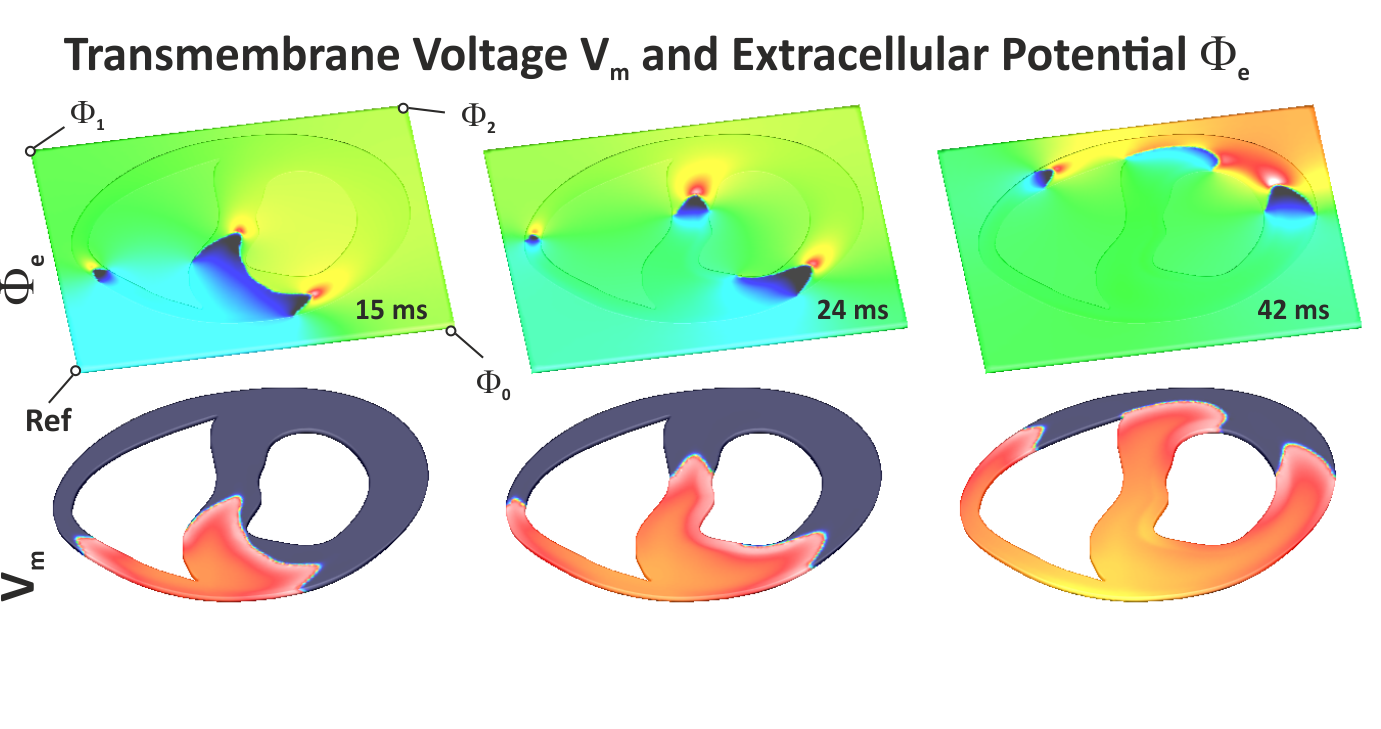
Fig. 30 Shown are extracellular potentials :math:’Phi_e’ and transmembrane voltage :math:’V_m’ induced by a :math:’S_1’ stimulus in the rabbit ventricular slice.
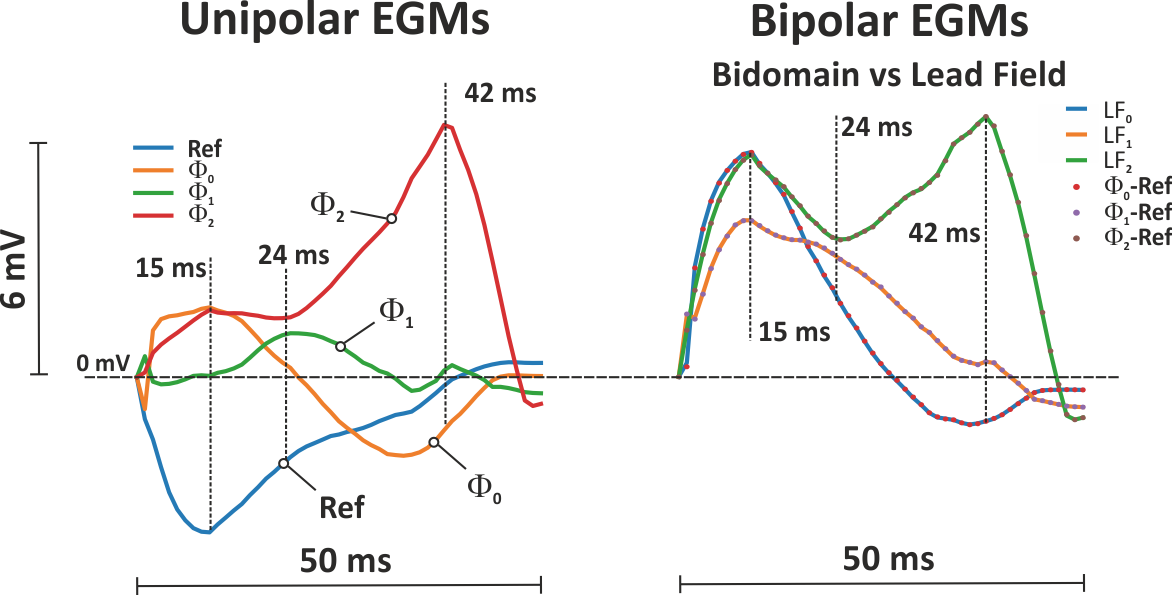
Fig. 31 Left panel: Shown are the extracellular potential traces recorded at the four corners of the bath. Right panel: Shown are bidomain traces (dotted lines) next to those computed using the lead field approach, showing that the lead field yields the exact bipolar bidomain EGMs.
Sinus activation
Wavefront propagation is initiated at the His bundle by the input parameter --purkStim His.
We use a normal His-Purkinje system (HPS) which is selected through --purkinje normal.
As we are only interested in the sinus activation we turn of any stimulation
which would trigger additional activations. This is achieved by setting --stimulus none.
./run_ep_v2.py --geometry 2C --purkStim His --purkinje normal --duration 50 \
--propagation R-E- --stimulus none --ID 2C_His_sinus_activation --visualize
Note
The 4C model in its current incarnation is only a baseline model for technical testing. Any capabilities in terms of more useful stimulation protocols must be added.