Basic usage of single cell tool bench - Limit cycle experiments
Module: tutorials.01_EP_single_cell.01_basic_bench.run
Section author: Gernot Plank <gernot.plank@medunigraz.at>
Modeling EP at the cellular level
This tutorial introduces the basic steps of setting up EP simulations in an isolated myocytes. A more detailed account on modeling EP at the cellular scale is given in the manual. For all experiments shown in this tutorial we use the single cell EP tool bench. Before starting we recommend therefore to look up the basic command line options in the bench section of the manual. In this introductory tutorial we focus on simple pacing protocols for finding a stable limit cycle. We model an isolated myocyte which we stimulate using an electrode for intracellular current injection. The setup is shown in Fig. 54.
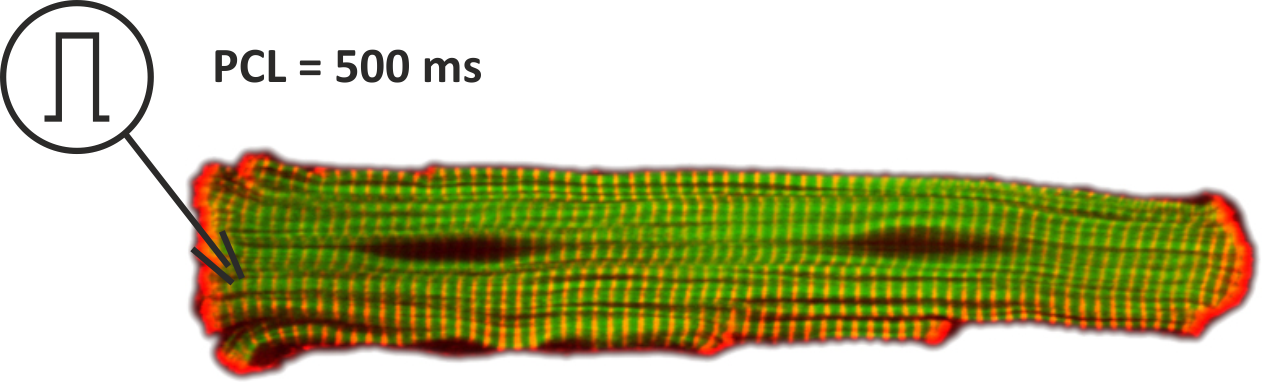
Fig. 54 Setup for single cell EP experiments. The myocyted is activated with a suprathreshold stimulus current to initiate action potentials at a pacing cycle lenght of 500 ms.
Experimental Parameters
The following parameters are exposed to steer the experiment:
--EP {TT2,GPB} pick human EP model (default is TT2)
--EP-par EP_PAR provide a parameter modification string (default is '')
--init INIT pick state variable initialization file (default is none)
--duration DURATION pick duration of experiment (default is 500 ms)
--bcl BCL pick basic cycle length (default is 500 ms)
--svRefH5 SVREFH5 pick a state variable h5 data set as reference for
comparison (default is none)
--curRefH5 CURREFH5 pick a current variable h5 data set as reference for
comparison (default is none)
--svfilter SVFILTER filter and show only pre-selected state variable
traces, if False, all traces are shownn (default is False)
--showCurrents SHOWCURRENTS
show current variable traces in addition to state
variables (default is False)
Each experiment stores the state of myocyte and myofilament in the current directory in a file to be used as initial state vector in subsequent experiments.
Limit cycle experiments
Background on the computaiton of limit cycles is found in the limit cycle section of the manual. Briefly, models of cellular dynamics are virtually never used with the values given for the initial state vector. Rather, cells are based to a given limit cycle to approximate the experimental conditions one is interested in. The state of the cell can be frozen at any given instant in time and this state can be reused then in subsequent simulations as initial state vector. In the following experiments the procedures for finding an initial state vector for a given model and a given basic cycle length are elucidated. To run the experiments of this tutorial change directories as follows:
cd ${TUTORIALS}/01_EP_single_cell/01_basic_bench
To see all the exposed parameters itemized above in experimental parameters section.
run
./run.py --help
Experiment exp01 (short pacing protocol)
We start with pacing the TT2 human myocycte model at a pacing cycle length of 500 ms for a duration of 5 seconds (5000 msecs).
./run.py --EP TT2 --duration 5000 --bcl 500 --ID exp01 --visualize --showCurrents True
To get an overview of all traces and currents of the model select the Check all option and
Replot in limpetGUI.
Experiment exp02 (limit cycle pacing protocol)
In experiment exp01 we observed that the state variables of the model did not stabilize at a stable limit cycle. We increase therefore the duration of our pacing protocol to 20 seconds (20000 msecs).
./run.py --EP TT2 --duration 20000 --bcl 500 --ID exp02 --visualize --showCurrents True
At the end of the pacing protocol the initial state vector is saved in the file exp02_TT2_bcl_500_ms_dur_20000_ms.sv.
-85.1192 # Vm
- # Lambda
- # delLambda
- # Tension
- # K_e
- # Na_e
- # Ca_e
0.00302465 # Iion
- # tension_component
TT2
0.13553 # Ca_i
3.90357 # CaSR
0.000390854 # CaSS
0.904553 # R_
2.8959e-07 # O
7.96934 # Na_i
137.474 # K_i
0.00176341 # M
0.740048 # H
0.668894 # J
0.0173138 # Xr1
0.469827 # Xr2
0.0147276 # Xs
2.47707e-08 # R
0.999995 # S
3.43115e-05 # D
0.72632 # F
0.958372 # F2
0.995453 # FCaSS
Ionic model parameter modification experiments
Model behavior can be modified by providing parameter modification string through --imp-par.
Modifications strings are of the following form:
param[+|-|/|*|=][+|-]###[.[###][e|E[-|+]###][\%]
where param is the name of the paramter one wants to modify
such as, for instance, the peak conductivity of the sodium channel, GNa.
Not all parameters of ionic models are exposed for modification,
but a list of those which are can be retrieved for each model using imp-info.
As an example, let’s assume we are interested in specializing a generic human ventricular myocyte
based on the ten Tusscher-Panfilov model to match the cellular dynamics of myocytes
as they are found in the halo around an infract, i.e.in the peri-infarct zone (PZ).
In the PZ various current are downregulated.
Patch-clamp studies of cells harvested from the peri-infract zone have reported
a 62% reduction in peak sodium current, 69% reduction in L-type calcium current
and a reduction of 70% and 80% in potassium currents 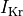 and
and 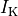 , respectively.
In the model these modifcations are implemented by altering the peak conductivities of the respective currents as follows:
, respectively.
In the model these modifcations are implemented by altering the peak conductivities of the respective currents as follows:
# both modification strings yield the same results
--imp-par="GNa-62%,GCaL-69%,Gkr-70%,GK1-80%"
# or
--imp-par="GNa*0.38,GCaL*0.31,Gkr*0.3,GK1*0.2"
In this experiment EP model parameters strings can be passed in using the --EP-par
which expects parameter strings as input.
The results of such modifications are illustrated in the following experiments.
Experiment exp03 (short pacing with modified myocyte)
In this experiment we modify the behavior of a TT2 myocyte to match experimental observations
of myocytes harvested from peri-infarct zones in humans.
Initially, we simulated one cycle only to observe the effect of parameter modifications
relative to the baseline TT2 model.
The referece traces are passed in through the --svRefH5 input
which takes the state variable and current traces stored in the experiment exp01.
./run.py --EP TT2 --duration 5000 --bcl 500 --ID exp03 --EP-par "GNa-62%,GCaL-69%,Gkr-70%,GK1-80%" \
--visualize --showCurrents True --svRefH5 exp01_TT2_traces.h5 --curRefH5 exp01_TT2_current_traces.h5
Make sure that the parameter modification string is correctly interpreted by bench by inspecting the output log:
./run.py --EP TT2 --duration 500 --bcl 5000 --ID test --EP-par "GNa-62%,GCaL-69%,Gkr-70%,GK1-80%"
...
Ionic model: TT2
GNa modifier: -62% value: 5.63844
GCaL modifier: -69% value: 1.2338e-05
Gkr modifier: -70% value: 0.0459
GK1 modifier: -80% value: 1.081
...
Experiment exp04 (limit cycle pacing with modified myocyte)
We repeat exp03 as a limit cycle experiment now by pacing for 20 seconds (20000 msecs). State variable and current traces stored in the baseline experiment exp02 are used as a reference.
./run.py --EP TT2 --duration 20000 --bcl 500 --ID exp04 --EP-par "GNa-62%,GCaL-69%,Gkr-70%,GK1-80%" \
--visualize --showCurrents True --svRefH5 exp02_TT2_traces.h5 --curRefH5 exp02_TT2_current_traces.h5
Visual comparison of the traces reveals that Calcium-cycling in the peri-infarct myocyte shows alternans. The difference in initial state vectors between baseline and peri-infarct myocyte stored at the end of the pacing protocol can be inspected with the meld or any other diff programmm of your choice.
meld exp02_TT2_bcl_500_ms_dur_20000_ms.sv exp04_TT2_bcl_500_ms_dur_20000_ms.sv
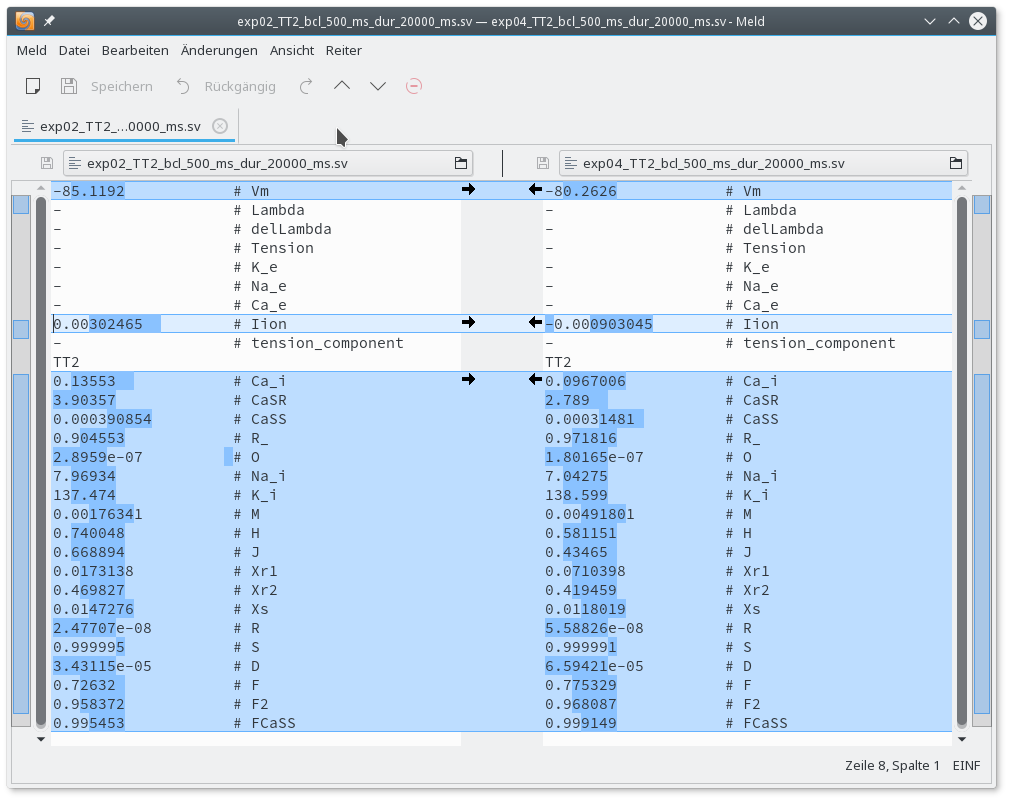
Fig. 55 Comparing initial state vectors of baseline and peri-infarct TT2 human ventricular myocyte model after 20 seconds of pacing at a basic cycle length of 500 ms.